Unearthing the origins of developmental heterogeneity in human acute myeloid leukemia
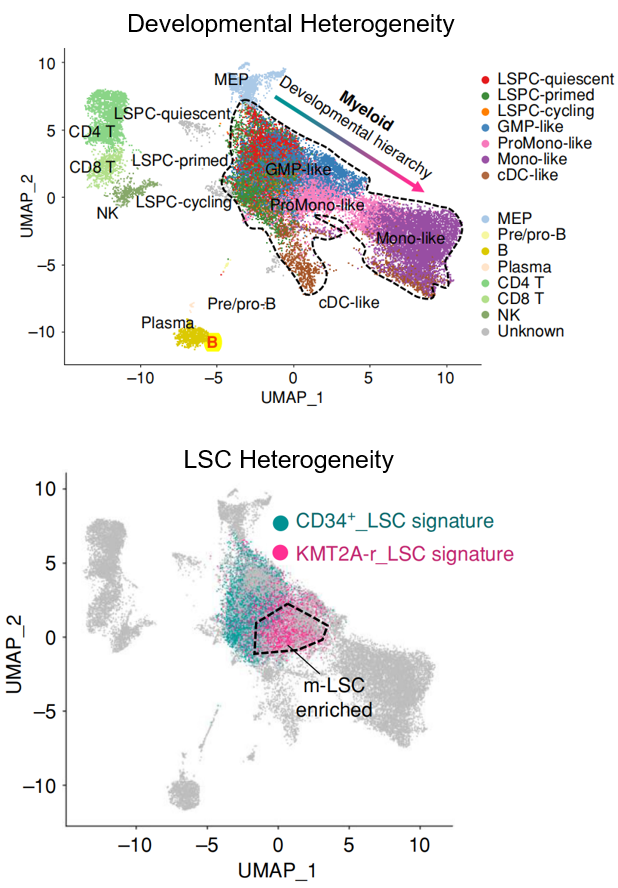
Developmental heterogeneity describes the diverse range of cell states present within the developmental hierarchy of a cancer tissue. It is increasingly recognized as a critical feature across a spectrum of malignancies, spanning solid tumors and hematological cancers, with particular significance in acute myeloid leukemia (AML). Distinct from genetic heterogeneity, developmental heterogeneity represents a key mechanism of therapeutic resistance, posing a significant challenge to novel targeted therapies directed at non-mutation-driven vulnerabilities. While genomic sequencing has greatly advanced our understanding of genetic heterogeneity, the origins and effective targeting strategies for developmental heterogeneity in human cancer remain largely uncharted. Recent work from Dr. Pei (Cancer Discov. 2023, ESI Top 1% Highly Cited Paper) has begun to illuminate these origins in AML, revealing a novel leukemia stem cell (LSC) subtype characterized by monocytic features. This study identified a previously unacknowledged population of monocytic LSCs coexisting with conventional primitive LSCs in a subset of AML patients, termed Multi-MMP AML. Notably, while primitive LSCs demonstrated sensitivity to BCL2 inhibition by venetoclax, the co-existing monocytic LSCs exhibited intrinsic resistance and were responsible for relapse following venetoclax-based therapy. This discovery underscores the profound complexity of LSC heterogeneity and its direct bearing on treatment outcomes, prompting fundamental questions about the fundamental nature of AML stem cells and their contribution to developmental heterogeneity and therapeutic resistance. Ongoing research in the Pei Lab is dedicated to comprehensively characterizing LSC heterogeneity in human AML, elucidating its biological links to genetic mutations, and ultimately defining its clinical implications for therapeutic response and resistance in AML.
Steering the path of clonal evolution in acute myeloid leukemia
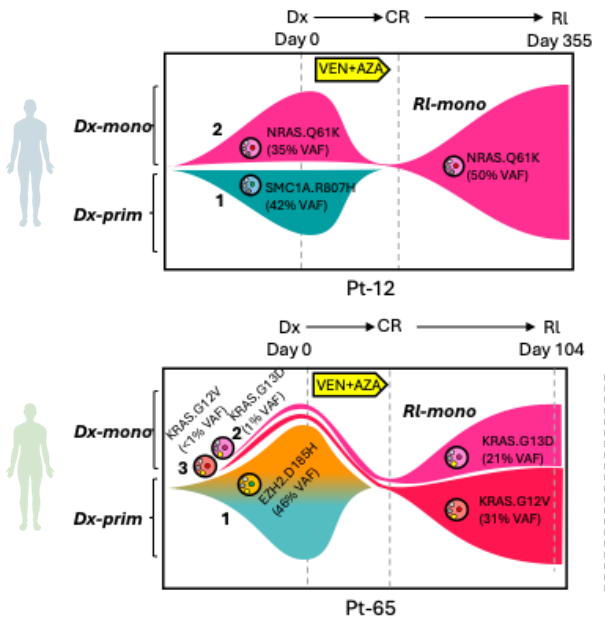
Acute myeloid leukemia (AML) is characterized by remarkable genetic and phenotypic heterogeneity. Genetically distinct subclones, existing in varying cellular states, undergo a dynamic evolutionary process under the selective pressure of therapeutic treatments, a phenomenon known as clonal evolution. At diagnosis, AML presents not as a uniform disease, but rather as a complex collection of leukemic subclones. Therapeutic interventions, such as chemotherapy, BCL2 or menin inhibitors, act as selective pressures, effectively eradicating sensitive clones while unfortunately allowing pre-existing or newly arising resistant subclones to survive and expand. This process of clonal evolution is a major obstacle to effective treatment and a primary driver of relapse and therapy resistance in AML. Therefore, deciphering the intricate patterns of clonal evolution under treatment pressure is paramount. By carefully studying how AML subclones adapt and reshape their clonal landscape in response to therapy, we can identify the mechanisms driving resistance and pinpoint vulnerable nodes in the evolutionary trajectory. Dr. PeiÔÇÖs previous work revealed for the first time that RAS-driven monocytic subclones emerged as the predominant drivers of relapse in response to Venetoclax-based therapy targeting BCL2 (Cancer Discov. 2020, AACR Most Cited Article Series, ESI Top 1% Highly Cited Article), a phenomenon that is broadly recognized in the AML clinic. Ongoing work in the Pei Lab continues to dissect the clonal evolution patterns of AML when challenged by various forms of molecular and cellular therapies. Ultimately, the goal is to develop strategies that not only target the dominant clone at diagnosis but also anticipate and effectively steer the path of clonal evolution, leading to more durable remissions and improved outcomes for patients with this aggressive leukemia.
Fighting the war between immune system and acute myeloid leukemia cells
Acute myeloid leukemia (AML) largely belongs to a group of immune-cold tumors. Most of AML cases have shown overwhelming de novo resistance to immune checkpoint blockade (ICB), and only a portion of immune-infiltrated AML cases responded to flotetuzumab immunotherapy. However, the observation that the chronic immunosuppression after organ transplantation greatly increased AML incidence suggested the existence of endogenous immunosurveillance against AML albeit it probably sustain at a restrained level. On the other hand, allogeneic hematopoietic stem cell transplantation (allo-HCT) is an established therapy with curative potential in AML. Nevertheless, post-transplant relapse is common and associated with poor prognosis. The occurrence of relapse after initially successful allo-HCT indicates that the donor immune system is first able to control the leukemia, which at a later stage develops evasion strategies to escape from immune surveillance. Thus, unleashing repressed immunosurveillance against AML holds great potential to improve AML treatment.
Previous work from Dr. Guo has demonstrated that abnormal IL-36 production is an essential feature of AML cells (Guo et al. Science Advances, 2021). IL-36 directly activates inflammatory monocytes (IMs) in bone marrow, which then precludes clearance of leukemia mediated by CD8+ T cells. While sparing IMs, chemotherapeutic agents stimulate IL-36 production from residual AML cells, thereby enabling the persistence of this immunosuppressive IL-36-IM axis after chemotherapy. Furthermore, IM depletion by trabectedin, with chemotherapy and PD-1 blockade, can synergistically restrict AML progression and relapse. Latest work from Dr. Guo found that AML cells activate CD36-mediated innate immune signaling to suppress T cell activity by transporting lipids (Guo et al. Cell Reports Medicine, 2024).High-fat-diet or decitabine treatment dampens the therapeutic effect by hijacking CD36 signaling. Targeting the CD36 immunosuppressive pathway with statins improves the efficacy of decitabine therapy in AML.
Now, we plan to identify new molecular mechanisms of immune escape in AML by unbiased genetic screens using CRISPR/Cas9. We focus on immune evasion strategies include downregulation of HLA, upregulation of inhibitory or downregulation of activating receptors, exhausted and dysfunctional immune effector cells. We hope that our study can provide rational treatment to help the immune system of patients with AML to win the war.
Implementing precision medicine from ÔÇ£bed to bench back to bedÔÇØ
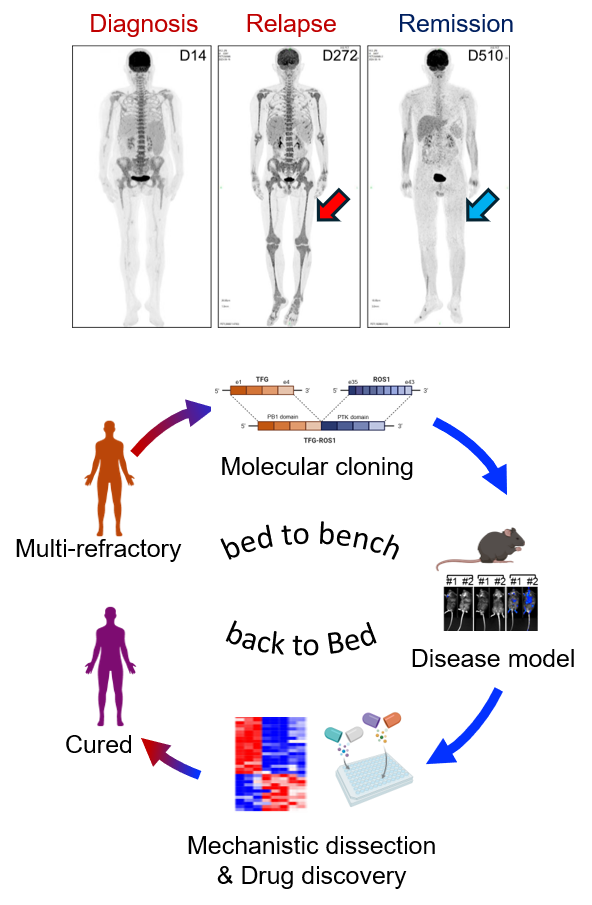
ÔÇ£Bed to bench back to bedÔÇØ exemplifies the ideal translational research loop, crucial for combating life-threatening malignancies like leukemia. Precision medicine offers transformative potential in cancer research, particularly for patients with hematological malignancies. The paradigm shift began with the discovery of oncogenic fusion proteins such as BCR-ABL in chronic myeloid leukemia and PML-RARA in acute promyelocytic leukemia. These breakthroughs paved the way for remarkably effective targeted therapies like imatinib and all-trans retinoic acid (ATRA) plus arsenic trioxide, respectively, transforming these once-deadly diseases into largely curable conditions. However, many rarer oncogenic fusions in hematological malignancies still await discovery and targeted intervention. Compellingly, Dr. Shaoqi Zhang, previously a co-mentored graduate student in the Pei Lab and now a physician-scientist in hematology, presented a remarkable example of precision medicine at the 2024 ASH meeting in San Diego. Through bioinformatic analysis and molecular cloning, Shaoqi identified a rare but recurrent TFG-ROS1 fusion in a multi-refractory leukemia patient who had progressed through chemotherapy, venetoclax treatment, and bone marrow transplant. She demonstrated its oncogenic driver role in both in vitro BaF3 models and in vivo syngeneic mouse models, via MEK/ERK activation and its exquisite sensitivity to ALK/ROS1 inhibitors like Ceritinib. Remarkably, this basic discovery enabled the successful translational application of Ceritinib in the clinic, resulting in complete remission in the patient for more than twelve months. This case powerfully showcases the potential of bench-to-bedside translational research. Given that cancer is fundamentally a genetic disease, ongoing work in the Pei Lab will continue to comprehensively identify and characterize rare oncogenic drivers in leukemia, accelerate the development of matched targeted therapies, and implement robust diagnostic platforms to deliver personalized and precision-based treatments, ultimately improving outcomes for more leukemia patients.
Attacking cellular vulnerabilities in human acute myeloid leukemia stem cells
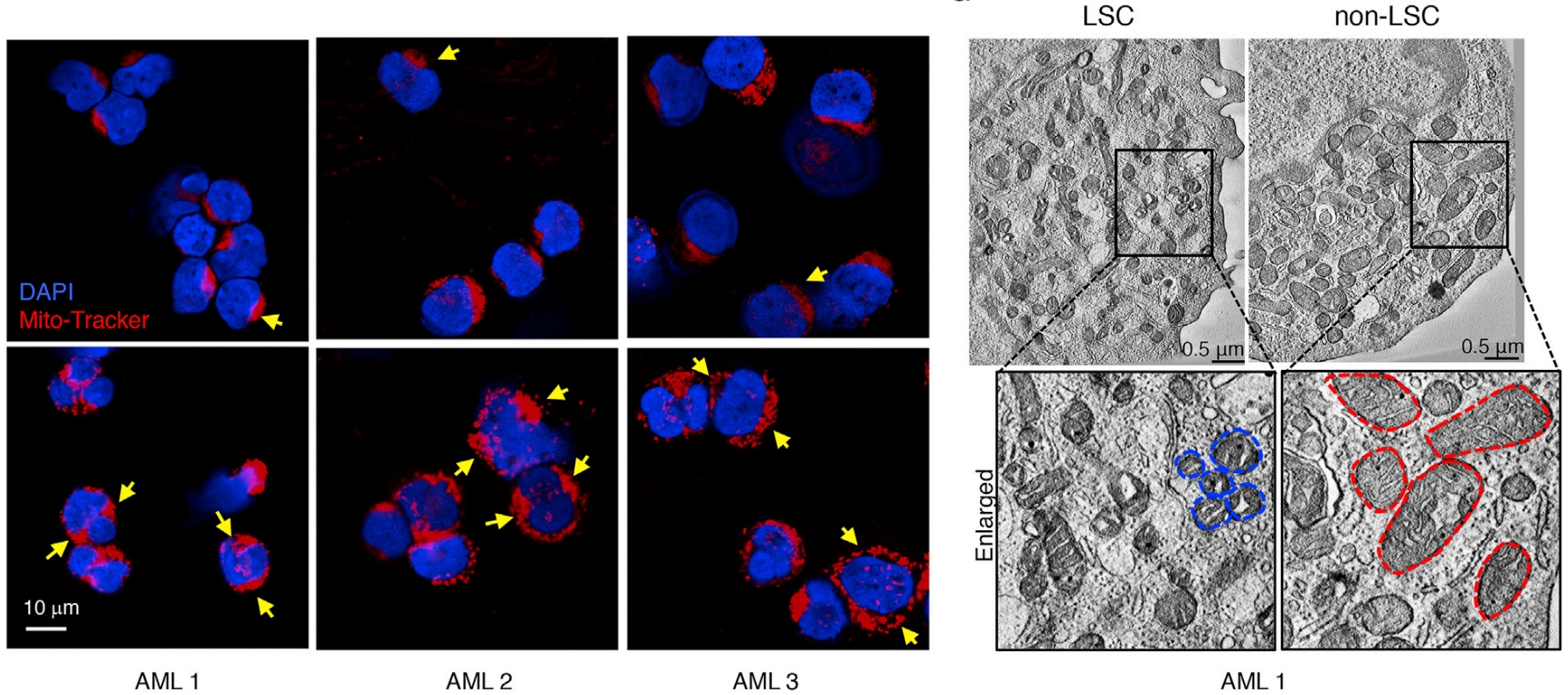
AML is a challenging cancer to treat due to the presence of Leukemia Stem Cells (LSCs), which drive disease progression and relapse. These LSCs are distinct from bulk tumor cells in many aspects, including their cellular biology. Mitochondria, key cellular organelles, have attracted significant attention in the AML field. Studies have revealed that multiple mitochondrial processesÔÇösuch as translation, transcription, energy metabolism, dynamics, mitophagy, resident protease activity, and their connection to epigenetics through metabolic intermediatesÔÇörepresent plausible therapeutic targets for treating AML and other cancers. However, how LSCs uniquely regulate mitochondrial biology to sustain their self-renewal potential and how different AML subtypes adapt their mitochondrial biology to cooperate with oncogenic drivers remain poorly understood. A previous study by Dr. Pei investigated the role of mitochondrial biology, specifically AMPK/FIS1-mediated mitophagy, in the self-renewal of AML LSCs (Cell Stem Cell, 2018). This study addressed how mitophagy contributes to maintaining a low but functional state of the mitochondrial network, sufficient to meet the unique metabolic and redox needs of LSCs, and whether targeting this pathway could selectively eradicate LSCs. Beyond mitophagy, oxidative phosphorylation (OXPHOS) is another vulnerability of LSC mitochondria. Work from Dr. PeiÔÇÖs previous labÔÇöDr. Craig JordanÔÇÖs groupÔÇörevealed that venetoclax, a milestone FDA-approved AML therapy targeting mitochondrial protein BCL2, used in combination with azacitidine, eradicates human LSCs by targeting mitochondrial OXPHOS (Nature Medicine, 2018), a unique dependency of human LSCs previously also identified by JordanÔÇÖs group through metabolic studies (Cell Stem Cell, 2013). In a recent collaboration with Dr. Craig JordanÔÇÖs lab, we discovered that mitochondrial calcium metabolism plays a key role in venetoclax resistance, and targeting the mitochondrial calcium uniporter (MCU) with the conventional therapeutic agent mitoxantrone can alleviate this resistance in human LSCs (Cancer Discovery, 2024). Ongoing work in the Pei Lab continues to explore the fascinating mitochondrial and other cellular biology of human LSCs, with a focus on discovering novel therapeutic targets.
Illuminating the molecular dependencies of human leukemia stem cells
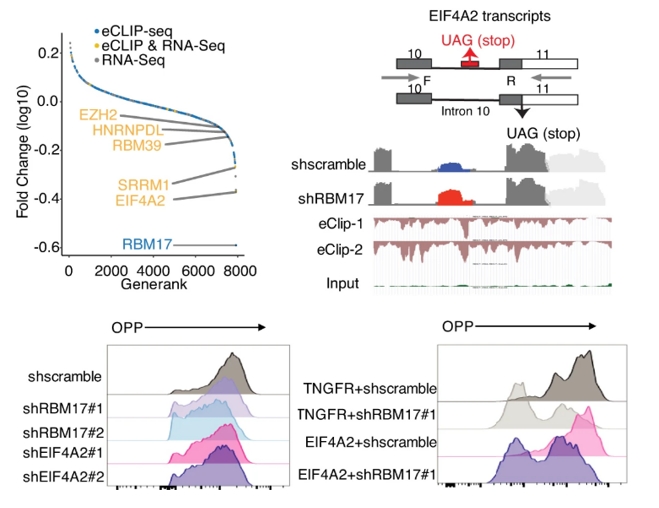
Illuminating the molecular dependencies of human leukemia stem cells (LSCs) is paramount to overcoming the persistent challenge of relapse in acute myeloid leukemia (AML). Current research has identified several potential molecular targets within AML and LSCs, including anti-apoptotic proteins like BCL2, signaling pathways such as FLT3 and RAS, and metabolic dependencies like mitochondrial oxidative phosphorylation. Our latest work revealed the purine metabolism is a unique vulnerability of monocytic LSCs (Cancer Discov. 2023, ESI Top 1% Most Cited Article). However, crucial questions remain: how do LSCs uniquely differ from normal hematopoietic stem cells and bulk leukemia cells in their molecular wiring? What are the non-oncogene addiction dependencies crucial for LSC survival and self-renewal across diverse AML subtypes? Furthermore, how can we effectively translate these molecular insights into novel therapeutic strategies that specifically eradicate LSCs while sparing normal stem cells? Previous work from Dr. Lina Liu, now a research professor in the Pei Lab,explored molecular dependency of AML LSCs at the level of postÔÇôtranscriptional control, where RNAÔÇÉbinding proteins (RBPs) function as core effectors. Armed with multiomic eCLIP-seq and RNA-seq analyses, Lina identified an RNA-binding protein called RBM17 that preferentially marks and sustains LSC and directly correlates with shorten patient survival in AML. Genetic targeting of RBM17 consequently results in inclusion of poison exons and production of nonsense-mediated decay (NMD)-sensitive transcripts for pro-leukemic factors (RBM39, HNRNPDL and EZH2) and the translation initiation factor, EIF4A2 (Nature Communications, 2022). Future research should focus on comprehensive multi-omic profiling of LSCs across AML subtypes, functional validation of novel dependencies, and the development of innovative targeted therapies, including molecularly targeted agents and immunotherapies, ultimately aimed at dismantling the LSC molecular machinery and achieving lasting cures in AML.
Perturbing metabolic plasticity for therapeutic opportunities
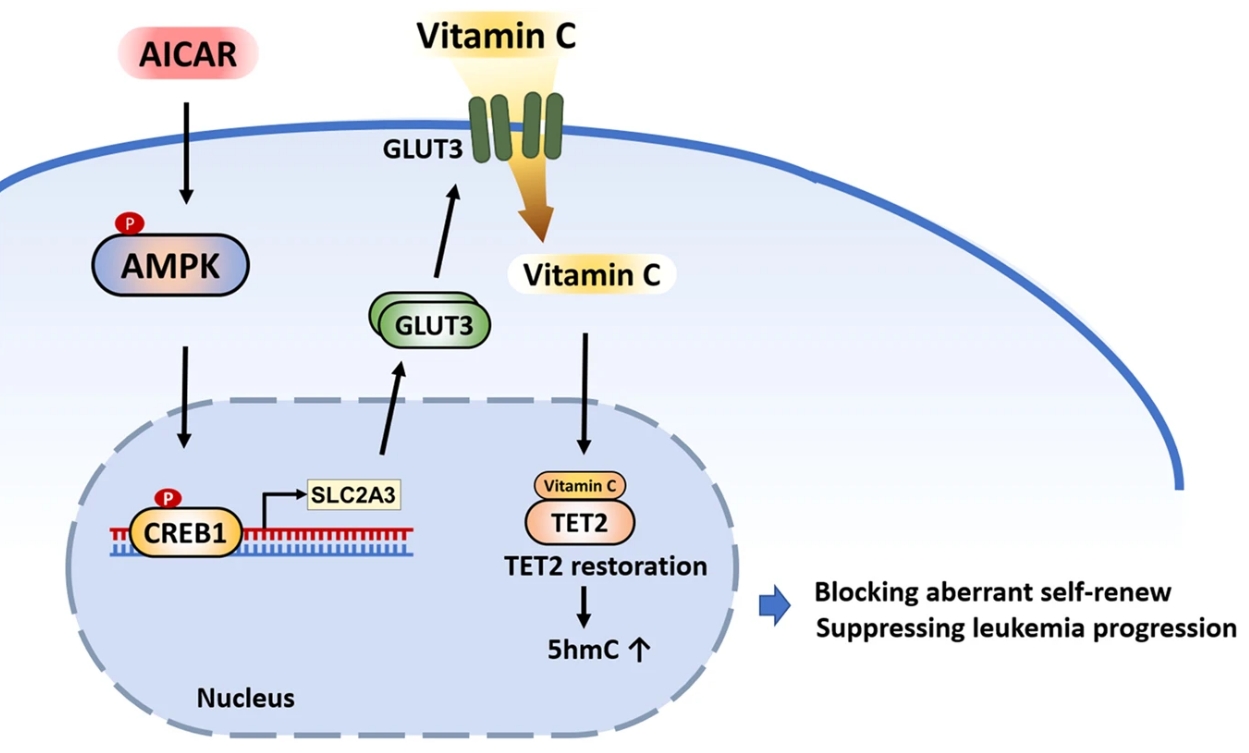
Metabolic plasticity, the capacity of cancer cells to dynamically adapt their metabolic pathways, is crucial for oncogenesis, tumor progression, and evasion of therapeutic interventions. It is increasingly recognized as a critical determinant in hematological malignancies, particularly acute myeloid leukemia (AML). This inherent metabolic flexibility empowers AML cells, and especially leukemia stem cells (LSCs), to withstand nutrient deprivation and therapeutic assaults, significantly contributing to chemoresistance and, notably, resistance to targeted therapies such as venetoclax. Building upon prior research, Dr. Jun Liu, now a postdoctoral fellow in the Pei Lab, investigated strategies to perturb metabolic plasticity for therapeutic advantage (Leukemia, 2023). His study addressed the challenge of limited vitamin C efficacy in TET2-mutated AML, revealing that insufficient expression of the vitamin C transporter GLUT3 restricts TET2 restoration and diminishes therapeutic response. However, pharmacological upregulation of GLUT3 using AICAR effectively overcame this limitation, enhancing TET2 activity and dramatically potentiating the anti-leukemic effects of vitamin C in preclinical models. Dr. Liu is now expanding his investigations to comprehensively understand global metabolic diversity in human AML. Ongoing research, spearheaded by Dr. Liu in the Pei Lab, aims to comprehensively map the diverse metabolic adaptations employed by AML cells and LSCs, dissect the precise role of metabolic plasticity in driving disease evolution, and develop innovative therapeutic strategies to effectively perturb this plasticity, ultimately improving treatment outcomes and overcoming drug resistance in AML.
